Research | Open Access | Volume 8 (3): Article 56 | Published: 24 Jul 2025
Occurrence of extended spectrum beta-lactamase-producing enterobacteriaceae at Edward Francis Small Teaching Hospital, Banjul, The Gambia 2022: A hospital-based study
Menu, Tables and Figures
Navigate this article
Tables
| Characteristics | Frequency | Percentage |
|---|---|---|
| Sex | ||
| Male | 79 | 45.66 |
| Female | 94 | 54.34 |
| Age Group | ||
| ≤10 | 39 | 22.54 |
| 11–20 | 19 | 10.98 |
| 21–30 | 52 | 30.06 |
| 31–40 | 19 | 10.98 |
| 41–50 | 20 | 11.56 |
| 51–60 | 17 | 9.83 |
| 61–70 | 4 | 2.31 |
| 71–80 | 2 | 1.16 |
| ≥81 | 1 | 0.58 |
| Total | 173 | 100 |
| Organisms | Female | Male | Total |
|---|---|---|---|
| Citrobacter spp | 1 | 0 | 1 |
| Enterobacter cloacae | 1 | 5 | 5 |
| E. coli | 41 | 18 | 59 |
| Enterobacter aerogenes | 1 | 0 | 1 |
| Escherichia vulneris | 0 | 1 | 1 |
| Pseudomonas fluorescence | 1 | 1 | 2 |
| Klebsiella oxytocka | 1 | 0 | 1 |
| Klebsiella pneumonia | 21 | 12 | 33 |
| Proteus spp. | 8 | 7 | 15 |
| Providence rettgeri | 0 | 1 | 1 |
| Pseudomonas luteola | 0 | 1 | 1 |
| Pseudomonas spp | 8 | 10 | 18 |
| Streptococcus pyogenes | 0 | 1 | 1 |
| Staphylococcus aureus | 9 | 20 | 29 |
| Serratia odrfera | 1 | 0 | 1 |
| Streptococcus pneumonia | 0 | 2 | 2 |
| Acinetobacter baumanii | 1 | 1 | 2 |
| Total | 94 | 79 | 173 |
| Bacteria | Positive | Negative | Total | ||
|---|---|---|---|---|---|
| Freq | % | Freq | % | ||
| E. coli | 21 | 60 | 38 | 40 | 59 |
| Klebsiella pneumoniae | 10 | 28.57 | 23 | 71.43 | 33 |
| Endobacteria cloacae | 2 | 5.71 | 3 | 94.25 | 5 |
| Klebsiella oxytoca | 1 | 2.85 | 0 | 96.88 | 1 |
| Providencia rettgeri | 1 | 2.85 | 0 | 0.00 | 1 |
| Proteus Spp | 0 | 0.00 | 15 | 100 | 15 |
| Fluorescence pseudomonas | 0 | 0.00 | 2 | 100 | 2 |
| Acinetobacter Baumannii | 0 | 0.00 | 2 | 100 | 2 |
| Citrobacter spp | 0 | 0.00 | 1 | 100 | 1 |
| Enterobacter Aerogenes | 0 | 0.00 | 1 | 100 | 1 |
| Escherichia vulneris | 0 | 0.00 | 1 | 100 | 1 |
| Pseudomonas utteola | 0 | 0.00 | 1 | 100 | 1 |
| Serratia orodifera | 0 | 0.00 | 1 | 100 | 1 |
| Total | 35 | 28.46 | 88 | 123 | |
Figures
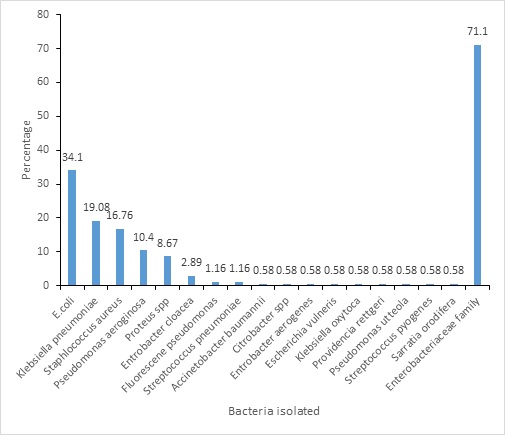
Figure 1: Frequency distribution of different bacterial isolates Bacteria Isolated by Sex

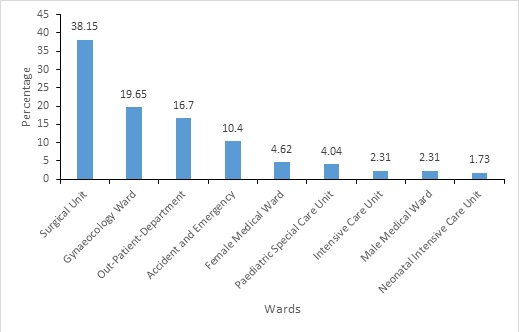
Figure 2: Frequency distribution of organisms by ward

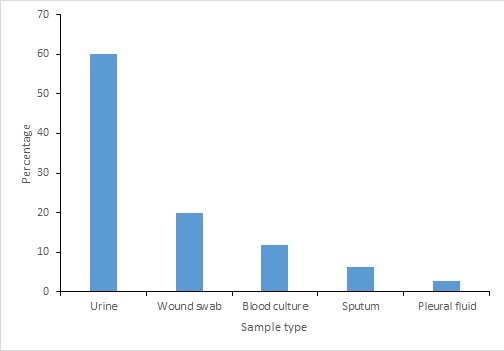
Figure 3: Distribution of ESBL Positive isolates by sample type

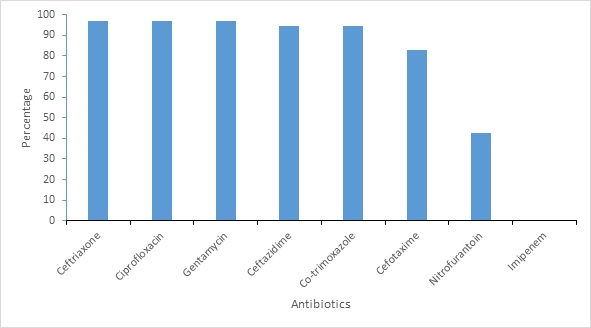
Figure 4: Antimicrobial resistance pattern of ESBL producing organisms

Keywords
- ESBL
- antibiotic
- resistant
- Enterobacteriaceae
- The Gambia
Ebrima Barrow1,2,&, Abou Kebbeh2,3, Haddy Bah1, Abdoulie Badjan1, Sainey Ceesay3, Kalipha Sanneh Darboe3, Baba Fofana2,4, Peter Adewuyi2
1Edward Francis Small Teaching Hospital, Microbiology Laboratory, Ministry of Health, Banjul, the Gambia, 2Gambia Field Epidemiology and Laboratory Training Program, Banjul, The Gambia, 3National Public Health Laboratories, Ministry of Health, Banjul, The Gambia, 4PhD candidate, University of Lahore, Lahore, Pakistan
&Corresponding author: Ebrima Barrow, Edward Francis Small Teaching Hospital, Microbiology Laboratory, Ministry of Health, Banjul, the Gambia. Email: ebrimabarrows@yahoo.com
Received: 30 May 2024, Accepted: 21 Jul 2025, Published: 24 Jul 2025
Domain: Antimicrobial Resistance
Keywords: ESBL, antibiotic, resistant, Enterobacteriaceae, The Gambia
©Ebrima Barrow et al Journal of Interventional Epidemiology and Public Health (ISSN: 2664-2824). This is an Open Access article distributed under the terms of the Creative Commons Attribution International 4.0 License (https://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Cite this article: Ebrima Barrow et al Occurrence of extended spectrum beta-lactamase-producing enterobacteriaceae at Edward Francis Small Teaching Hospital, Banjul, The Gambia 2022: A hospital-based study. Journal of Interventional Epidemiology and Public Health 2025;8:56. https://doi.org/10.37432/jieph.2025.8.3.174
Abstract
Introduction: Extended spectrum beta-lactamases producer enterobacteriaceae (ESBL_PE) are increasing in both hospital and community settings posing a major public health problem worldwide. In The Gambia, published data on the occurrence of ESBL-PE in local settings is limited. We determined the occurrence of ESBL-PE from clinical samples at the main tertiary hospital, Edward Francis Small Teaching Hospital (EFSTH), The Gambia.
Methods: From December 2021 to August 2022, 173 banked clinical isolates were analysed using standard bacteriological methods at the EFSTH microbiology laboratory. Isolates identification was conducted using biochemical tests. Antibiotic susceptibility was conducted using the disk diffusion method according to Clinical and Laboratory Standard Institute (CLSI) guidelines. Phenotypic ESBL- PE was confirmed using double-disk synergy methods. Data on demographic characteristics, ward and sample type was collected from laboratory registers and request forms. Data was analysed descriptively using Epi info 7.5.
Results: A total of 173 single clinical isolates were analysed. ESBL-PE frequency was 35 (28.46%) among the 123 Enterobacteriaceae isolates identified. Majority of the ESBL PE isolates were from gynaecology ward 14(40%), followed by the surgical ward, 11 (31.43%). Escherichia coli was dominant 21 (60%) followed by Klebsiella pneumoniae 10 (28.57%). A high resistance rate was observed against Cephalosporins [(Ceftazidime 34 (97.14%), Cefotaxime 29(82.9%), Ceftriaxone 33 (94.29%)], Cotrimoxazole 33 (94.29%) and Gentamycin 32 (91.43%). A low resistance rate of ESBL-producers against Nitrofurantoin 9 (42.9) was observed. However, all the ESBL isolates were sensitive to Imipenem antibiotics. Multi-drug-resistant isolates were more prevalent among the ESBL_PE 33 (94.29%) than non-producers 2 (5.71%) (p = <0.0001)
Conclusion: Our study shows ESBL-producing Enterobacteriaceae from the banked clinical isolates, with Escherichia coli and Klebsiella pneumoniae as the predominant organisms. ESBL-producing organisms show a high resistance rate to cephalosporins. The hospital management should implement infection prevention and control measures within gynaecology and surgical wards – the most affected wards to prevent the spread of ESBL-PE.
Keywords: ESBL, antibiotic, resistant, Enterobacteriaceae, The Gambia
Introduction
Antimicrobial Resistance (AMR) is a major public health threat worldwide including The Gambia. AMR occurs when microbials develops mechanism to resist the effects of antimicrobials rendering them ineffective in treating infections. Enzymatic production is one of the mechanisms by which microbials develop resistance to antimicrobials [1]. Extended spectrum beta-lactamases (ESBLs) are enzymes produced by microorganisms that hydrolyze the ß-lactam ring of penicillin, first, second and third generation and higher cephalosporins as well as aztreonam [2] . The enzymes are commonly expressed by Enterobacteriaceae [3]. The occurrence of ESBLs among Enterobacteriaceae is predominantly observed in Klebsiella. Spp and Escherichia coli as well as other Enterobacteriaceae families such as Citrobacter. Spp, Salmonella. Spp, Proteus. Spp [3-6]. Infections caused by Extended spectrum beta-lactamases producer enterobacteriaceae (ESBL PE) are increasingly becoming difficult to treat which has led to use of more expensive broad-spectrum antibiotics [4]. This is because of misusing antibiotics. [[7]. The inappropriate usage of antibiotics both in healthcare and community settings has been cited as the contributing factors in the emergence of ESBL PE leading to AMR worldwide. Use of antibiotics without medical prescription, missed used and abused and [8], hospitalization, catheterization [3] are some of the factors contributing to the emergence and spread of ESBL-PE. Globally, ESBL-PE- pose a serious challenge to both clinical care and public health leading to prolonged hospitalization, higher mortality and economic loss [9]. In 2019, an estimate of 4.95 million deaths associated with bacterial AMR were reported worldwide [10]. The increasing occurrence of these ESBL-PE has been documented worldwide and varies from one country to another among different population and age-groups; 80.9% among hospitalized patients in Togo [11], in Gaza strip 51.6% ESBL isolates were detected among pediatric patients [12], 39.2% ESBL producing Escherichia coli and Klebsiella pneumoniae reported in Senegal [13], 5.0% was observed among food handlers in The Gambia [5]. In The Gambia, few studies have been conducted to elucidate the occurrence of ESBL-PE, notably the study conducted by Sanneh et.al was conducted on healthy carriers at community level [5]. According to Global Research on antimicrobial resistance (GRAM) 2019 report the number of AMR deaths in The Gambia is higher than deaths from maternal and neonatal disorders, HIV/AIDS and sexually transmitted infections, neoplasms, enteric infections, neglected tropical diseases and malaria. In 2019, The Gambia had 402 deaths attributable to AMR and 1800 associated death with AMR It is evident that there is limited information on the occurrence of ESBL-PE among patients at Edward Francis Small Teaching Hospital (EFSTH) in The Gambia. This study determined the prevalence of ESBL-PE at this hospital.Methods
Study Design
A retrospective cross-sectional study was conducted from September to December 2022 on bacterial isolates at the EFSTH microbiology laboratory. Isolates banked from December 2021 to August 2022 were used. The banked clinical bacterial isolates from various clinical samples: wound, blood, urine, ear swabs, pus, cerebrospinal fluid (CSF), sputum and pleural fluids were sub-cultured to determine ESBL-PE. Data on age, sex, ward and sample type were collected from registers and patient forms.
Study Setting
The Edward Francis Small Teaching Hospital (EFSTH) is the only tertiary teaching hospital and the main referral health facility in The Gambia. EFSTH is a 600-bed-capacity tertiary hospital located in the capital, Banjul. The hospital offers consultant and general services in the different clinical disciplines of Surgery, Internal Medicine, Laboratory, Pediatrics, Obstetrics & Gynaecology, Family Medicine and a recently introduced Cardiology. The laboratory department of EFSTH has the following specialities: microbiology, haematology, biochemistry, serology, blood bank, histology and parasitology. The microbiology laboratory department received clinical samples from different wards and mainly conducts bacteriological identification and sensitivity testing.
Sub-culture and Identification of Banked Bacterial Isolates
The isolates preserved on glycerol broth at –800C were retrieved using standard microbiology culture methods. Isolates were sub-cultured on blood agar, chocolate agar and MacConkey agar (Oxoid Ltd, Basingstoke, United Kingdom) and incubated between 18-24 hours at 370C. After the incubation, organisms were characterized by colony morphology, Gram stain and biochemical reactions [14]. Enterobacteriaceae were identified using commercial Analytical Profile Index 20E gallery (BioMeriuex, France) [11].
Antimicrobial Susceptibility Testing
Antimicrobial Susceptibility Testing (AST) was conducted using Kirby-Bauer’s disc diffusion method following standard guidelines of Clinical Laboratory Standards Institute (CLSI) [15]. A single well-isolated colonies from an overnight culture were selected, emulsify in 5ml sterile saline solution. The organism suspension turbidity was compared with a 0.5 McFarland standard. After preparation of the inoculum suspension, Muller-Hinton agar (Oxoid Ltd, Basingstoke, United Kingdom) was inoculated and antibiotics disc were aseptically placed [16]. Bacterial isolates were tested against the following antibiotics that are readily available: ampicillin (30 μg), piperacillin (100μ g), amoxicillin/clavulanic acid (20 μg), cefotaxime (30 μg), ceftazidime (30 μg), ceftriaxone (30 μg), imipenem, ciprofloxacin (30μg), gentamycin (30μg) and cotrimoxazole (30 μg).[6, 9]
Phenotypic ESBL Detection
Isolates that showed reduced inhibition zone size of ≤22mm with ceftazidime (30µg) and/or ≤27mm with cefotaxime (30µg) were considered as potential ESBL-producer and were selected for ESBL confirmation [14]. Enterobacteriaceae isolates were confirmed for phenotypic ESBL-producer based on a ≥ 5 mm increase in a zone diameter for either cefotaxime or ceftazidime antibiotics tested in combination with clavulanate versus the zone of inhibition of the antibiotics when tested alone [15]. An isolate is considered multidrug-resistant if it is resistant to three or more classes of antibiotics [16].
Quality Control
Standard Operating procedures were followed in the laboratory analysis. Sterility and performance were conducted for every new batch of prepared media. Standard reference strains of ATCC 25922 E. coli (ESBL negative) and K. pneumoniae ATCC 700603 (ESBL positive) were used to quality control for the purchased antibiotic disc [15].
Data management and analysis
Data was entered into a Microsoft Excel sheet, cleaned, coded and analysed using statistical software STATA (version 16.1). Measures of central tendency were used to summarise the continuous variables. Proportion of ESBL-PE was used to describe the frequency of categorical variables. We determine the statistical difference among MDR isolates for ESBL-producer and non-ESBL-producers at 95% confidence level. Analyzed data were presented as text, graphs and tables.
Ethical considerations
Ethical approval for this study was obtained from the ethical committee of Edward Francis small teaching hospital (protocol number EFSTH 2023 109).
Results
Background characteristics
A total of 173 isolates were included in the study, most of which were from female patients 94 (54%) with a mean age of 30 years (SD±16). Majority of isolates were from patients aged between 21-30 years 52 (30.06%) followed by those aged ≤10 years 39 (22.54%). Only one bacterium was isolated from patients aged ≥ 81 years (0.57%) (Table 1).
Bacterial Isolates from Clinical Samples
Of the 173 isolates from clinical specimens, 59 (34.10%) were Escherichia coli, 33 (19.07%) was Klebsiella pneumoniae, 29 (16.7%) were Staphylococcus aureus, 18 (10.40%) Pseudomonas aeruginosa, 15 (8.67%) Proteus spp, 5 (2.89%) Enterobacter cloacae, 2 (1.15%) Pseudomonas fluorescence, 2 (1.15%) streptococcus pneumonia, 2 (1.15%) Acinetobacter baumanii; Citrobacter spp , Enterobacter aerogenese, Escherichia vulneris, Klebsiella oxytoca, Providencia rettgeri, Pseudomonas utteola, Streptococcus pygonese and serratia orodifera each constituted 1 (0.57%). Overall, 123 (71.1%) were from the family of Enterbacteriaceae (Figure 1).
Bacteria Isolated by Sex
Over half of the 173 isolated bacteria were from female patients, 94 (54.3%). In which 69.49% (41/59) females were infected with E. coli and 63.63% (21/33) females had Klebsiella pneumonia. Staphylococcus aureus was mostly isolated among male patients 69.0% (20/29) (Table 2).
Bacterial Isolates from different wards
Of the 173 clinical isolates 66 (38.15%) were isolated from surgical wards, 34 (19.65%) from gynaecology ward, 29 (16.76%) from outpatient, 18 (10.40%) from accident and emergency (A/E), 8 (4.62%) from female medical ward, 7 (4.04%) from PSCU, 4 (2.31%) from intensive care unit (ICU) and male medical ward respectively and 3 (1.73%) from neonatal intensive care unit (NICU).
ESBL producing isolates by sample type
Among the 123 entrobacteriaceae isolates, 35 ESBL-positive 21 (60%) were urine isolates, 7 (20%) were wound isolates, 4 (11.76%) were blood culture isolates, 2 (6.25%) was sputum isolates and 1 (2.85%) were pleural fluid isolates (Figure 3).
ESBL-Producing Enterobacteriaceae Isolates
Among the 123 isolates of Enterobacteriaceae from clinical specimens 35 (28.45 %) were positive for ESBL. Majority were E. coli 21 (60.0 %) followed by Klebsiella pneumoniae 10 (28.57%), 2 (5.71%) was Endobacteria cloacae and 1(3.12%) were Klebsiella oxytoca and Providencia rettgeri, respectively (Table 3).
Antimicrobial Resistance Profile of ESBL-Producing Organism
All ESBL isolates are sensitive to Imipenem (35/35). All the ESBL-producing Enterobacteriaceae were Multi-drug Resistant (MDR) showing resistant to at least three different classes of antibiotics. High resistant was observed in Ceftriaxone 34 (97.1%), Ciprofloxacin 34 (97.1%), Gentamycin 34 (97.1%), Followed by Ceftazidime 33 (94.3%), Co-trimoxazole 33 (94.29%), Cefotaxime 29 (82.9%) and Nitrofurantoin 9 (42.9%) resistance (Figure 4).
Discussion
Extended-spectrum beta-lactamases (ESBL) causing infections are increasing in incidence both in hospital and community settings. This poses a major public health threat for the treatment of infectious diseases caused by ESBL PE. The findings from our study that used banked clinical isolates from EFSTH shows that occurrence of ESBL was 28.6%, a much higher proportion compared to the study done by Sanneh et.al on food handlers 2008 which was 5% [12]. However, Sanneh’s study was conducted on a heathier population while this study was on isolates of patients. Another study done in a similar hospital setting showed high incident of 26.6% from clinical isolates Click or tap here to enter text.[17]. This suggests the need for proper continuous surveillance of AMR in hospital settings. This is evident from our findings which showed that E. coli and K. pneumonia were the most common ESBL PE isolated, and a previous study showed these two pathogens were the most common pathogens causing hospital acquired infections [2]. This is further supported by a similar study done in Togo in 2019 which also reported ESBL producing E. coli and klebsiella [4].
Furthermore, this study revealed higher frequency of ESBL PE among patients with severe infections including urinary tract infections, wound infections and blood stream infections. However, ESBL PE have been linked to serious hospital acquired infection worldwide leading to prolong hospitalization, higher morbidity and mortality [18]. This could be due to poor infection control measures such as poor hand hygiene and poor patient isolation systems. Also drug supply constraints in developing countries frequently limit prescribers to the medications that are readily available contributing further to AMR.
Our study showed that imipenem is the most effective antibiotic against ESBL producing isolates with 100% sensitivity, which is in agreement with similar published data in Burkina Faso, which showed a susceptibility of 100% for imipenem [19]. Our study shows high resistance of 89% and above towards the following cephalosporins ceftriaxone, cefotaxime and ceftazidime, which is similar to the Gambia study done by Sanneh [2] with 100% resistance recorded for these same drugs. It also demonstrated a similarly high resistance to non-cephalosporins ciprofloxacin, gentamycin and co-trimoxazole. This high resistance could be due to prolonged hospitalisation, invasive medical devices and over exposure to third generation cephalosporins. The latter is supported by a study done by Chaw et al at EFSTH, where Ceftriaxone- an antibiotic from the WHO watch group that is frequently used as second-line therapy, was one of the most widely used antibiotics in the hospital with a greater density of use than ampicillin a first-line antibiotic [20]. The study shows that all the ESBL PE are MDR organisms as ESBL is plasmid mediated and is linked to co-resistance to other antibiotic classes. Misuse of antibiotics, self-medication, misdiagnosis and empirical treatment before laboratory findings, easy access to antibiotics at pharmacies and private clinics could be the contribution factors for the higher antimicrobial resistance [21]. Additionally, we noted (4%) imipenem resistance from non ESBL producing organisms.
Conclusion
This study revealed the presence of ESBL PE among the EFSTH clinical isolates, with E. coli and Klebsiella pneumoniae as the predominant organisms. Most isolates that produced ESBLs were from urine and wound isolates. ESBL PE showed a greater degree of resistance to various antibiotic classes in contrast to non-ESBL producers. Imipenem is a alternative for treating ESBL PE. The emergence of multidrug-resistant bacteria (MDR) and ESBL PE requires strengthen of clinical bacteriology research and enhancing laboratory diagnostic abilities to identify and monitor antibiotic resistance. We advise for routine infection prevention strategies such as hand hygiene and rational used of antibiotics, proper sterilization as control measures for the spread of ESBL PE. Our study findings establish the baseline information required for more extensive investigations incorporating the genetic characterization of resistant strains to determine the origin, varieties, and patterns of antimicrobial resistance at Edward Francis small Teaching Teaching Hospital.
What is already known about the topic
- ESBL are enzymes that catalyse the betalactam antibiotics and render them ineffective.
- Infections caused by ESBL PE cause prolonged hospitalisation, higher morbidity and morbidity.
What this study adds
- The occurrence of ESBL PE from clinical isolates at EFSTH and an antibiogram showing susceptibility pattern to the locally available antibiotics.
- The sample type with the highest number of ESBL PE isolates.
Authors´ contributions
Design and Conceptualization: EB, AK. Data Collection and Investigation: EB. Laboratory Investigation: EB, AB, BKF. Data Analysis: EB,AK. Writing Original Draft: EB, AK, SBFC, KSD, BKF. Writing, Review and Editing: HB and PA. All the authors reviewed and approved the final version.
| Characteristics | Frequency | Percentage |
|---|---|---|
| Sex | ||
| Male | 79 | 45.66 |
| Female | 94 | 54.34 |
| Age Group | ||
| ≤10 | 39 | 22.54 |
| 11–20 | 19 | 10.98 |
| 21–30 | 52 | 30.06 |
| 31–40 | 19 | 10.98 |
| 41–50 | 20 | 11.56 |
| 51–60 | 17 | 9.83 |
| 61–70 | 4 | 2.31 |
| 71–80 | 2 | 1.16 |
| ≥81 | 1 | 0.58 |
| Total | 173 | 100 |
| Organisms | Female | Male | Total |
|---|---|---|---|
| Citrobacter spp | 1 | 0 | 1 |
| Enterobacter cloacae | 1 | 5 | 5 |
| E. coli | 41 | 18 | 59 |
| Enterobacter aerogenes | 1 | 0 | 1 |
| Escherichia vulneris | 0 | 1 | 1 |
| Pseudomonas fluorescence | 1 | 1 | 2 |
| Klebsiella oxytocka | 1 | 0 | 1 |
| Klebsiella pneumonia | 21 | 12 | 33 |
| Proteus spp. | 8 | 7 | 15 |
| Providence rettgeri | 0 | 1 | 1 |
| Pseudomonas luteola | 0 | 1 | 1 |
| Pseudomonas spp | 8 | 10 | 18 |
| Streptococcus pyogenes | 0 | 1 | 1 |
| Staphylococcus aureus | 9 | 20 | 29 |
| Serratia odrfera | 1 | 0 | 1 |
| Streptococcus pneumonia | 0 | 2 | 2 |
| Acinetobacter baumanii | 1 | 1 | 2 |
| Total | 94 | 79 | 173 |
| Bacteria | Positive | Negative | Total | ||
|---|---|---|---|---|---|
| Freq | % | Freq | % | ||
| E. coli | 21 | 60 | 38 | 40 | 59 |
| Klebsiella pneumoniae | 10 | 28.57 | 23 | 71.43 | 33 |
| Endobacteria cloacae | 2 | 5.71 | 3 | 94.25 | 5 |
| Klebsiella oxytoca | 1 | 2.85 | 0 | 96.88 | 1 |
| Providencia rettgeri | 1 | 2.85 | 0 | 0.00 | 1 |
| Proteus Spp | 0 | 0.00 | 15 | 100 | 15 |
| Fluorescence pseudomonas | 0 | 0.00 | 2 | 100 | 2 |
| Acinetobacter Baumannii | 0 | 0.00 | 2 | 100 | 2 |
| Citrobacter spp | 0 | 0.00 | 1 | 100 | 1 |
| Enterobacter Aerogenes | 0 | 0.00 | 1 | 100 | 1 |
| Escherichia vulneris | 0 | 0.00 | 1 | 100 | 1 |
| Pseudomonas utteola | 0 | 0.00 | 1 | 100 | 1 |
| Serratia orodifera | 0 | 0.00 | 1 | 100 | 1 |
| Total | 35 | 28.46 | 88 | 123 | |




References
- Rawat D, Nair D. Extended-spectrum β-lactamases in Gram negative bacteria. J Glob Infect Dis. 2010;2(3):263-74. Available from: https://journals.lww.com/10.4103/0974-777X.6853. doi:10.4103/0974-777X.68531.
- Akpaka PE, Vaillant A, Wilson C, Jayaratne P. Extended spectrum beta-lactamase (ESBL) produced by Gram-negative bacteria in Trinidad and Tobago. Int J Microbiol. 2021;2021:5582755. Solanki MK, editor. Available from: https://www.hindawi.com/journals/ijmicro/2021/5582755/. doi:10.1155/2021/5582755.
- Sarojamma V, Ramakrishna V. Prevalence of ESBL-producing Klebsiella pneumoniae isolates in tertiary care hospital. ISRN Microbiol. 2011;2011:318348. Available from: https://www.hindawi.com/journals/isrn/2011/318348. doi:10.5402/2011/318348.
- Godonou AM, Lack F, Gbeasor-Komlanvi FA, Konlani L, Dossim S, Ameyapoh YA, Ekouevi K, Dagnra AY, Salou M. High faecal carriage of extended-spectrum beta-lactamase producing Enterobacteriaceae (ESBL-PE) among hospitalized patients at Sylvanus Olympio Teaching Hospital, Lomé, Togo in 2019. Afr J Clin Exp Microbiol. 2022;23(1):40-8. Available from: https://www.ajol.info/index.php/ajcem/article/view/220654. doi:10.4314/ajcem.v23i1.6.
- Hasan B. Antimicrobial resistance and production of extended spectrum beta-lactamases in Enterobacteriaceae from birds in Bangladesh [dissertation]. Uppsala: Acta Universitatis Upsaliensis; 2013 [cited 2025 Jul 21]. Available from: https://uu.diva-portal.org/smash/get/diva2:620961/FULLTEXT01.pdf.
- Sharba ZA, Farazdaq Rafeeq H. The incidence of extended spectrum β-lactamase enzymes and their connection to virulence genes in community-acquired urinary tract infection. Rev Bionatura. 2022;7(2):27. Available from: https://www.revistabionatura.com/2022.07.02.27.html. doi:10.21931/RB/2022.07.02.27.
- Institute for Health Metrics and Evaluation. The burden of antimicrobial resistance (AMR) in The Gambia [Internet]. Seattle (WA): University of Washington; 2019 [cited 2025 Jul 21]. Available from: https://www.healthdata.org/sites/default/files/files/policy_report/2019/AMR_The_Gambia_2019.pdf.
- Bezabih YM, Sabiiti W, Alamneh E, Bezabih A, Peterson GM, Bezabhe WM, Roujeinikova A. The global prevalence and trend of human intestinal carriage of ESBL-producing Escherichia coli in the community. J Antimicrob Chemother. 2021;76(1):22-9. Available from: https://academic.oup.com/jac/article/76/1/22/5919862. doi:10.1093/jac/dkaa399.
- Baziboroun M, Bayani M, Poormontaseri Z, Shokri M, Biazar T. Prevalence and antibiotic susceptibility pattern of extended spectrum beta lactamases producing Escherichia coli isolated from outpatients with urinary tract infections in Babol, Northern of Iran. Curr Issues Pharm Med Sci. 2018;31(2):61-4. Available from: https://czasopisma.umlub.pl/curipms/article/view/659. doi:10.1515/cipms-2018-0013.
- Lohani B, Thapa M, Sharma L, Adhikari H, Sah AK, Khanal AB, et al. Predominance of CTX-M type extended spectrum β-lactamase (ESBL) producers among clinical isolates of Enterobacteriaceae in a tertiary care hospital, Kathmandu, Nepal. Open Microbiol J. 2019;13:28-33. Available from: https://openmicrobiologyjournal.com/VOLUME/13/PAGE/28/FULLTEXT/. doi:10.2174/1874285801913010028.
- Sanneh B, Kebbeh A, Jallow HS, Camara Y, Mwamakamba LW, Ceesay IF, Barrow E, Sowe FO, Sambou SM, Baldeh I, Jallow A, Jorge Raul MA, Andremont A. Prevalence and risk factors for faecal carriage of extended spectrum β-lactamase producing Enterobacteriaceae among food handlers in lower basic schools in West Coast Region of The Gambia. PLoS One. 2018;13(8):e0200894. Available from: https://dx.plos.org/10.1371/journal.pone.0200894. doi:10.1371/journal.pone.0200894.
- Alio MF, Laouali B, Ali M, Hadiza IB, Ali K, Chaibou Y, Cheikna Z, Chaibou S, Alhousseini D, Ramatou S, Alfred ST, Nicolas B. Phenotypic detection of extended spectrum beta-lactamase in multidrug-resistant Escherichia coli from clinical isolates in Niamey, Niger. Afr J Microbiol Res. 2017;11(18):712-7. Available from: http://academicjournals.org/journal/AJMR/article-abstract/A9FFC3264229. doi:10.5897/AJMR2017.8535.
- Ghafourian S, Sekawi Z, Neela V, Khosravi A, Rahbar M, Sadeghifard N. Incidence of extended-spectrum beta-lactamase-producing Klebsiella pneumoniae in patients with urinary tract infection. Sao Paulo Med J. 2012;130(1):37-43. Available from: http://www.scielo.br/scielo.php?script=sci_arttext&pid=S1516-31802012000100007&lng=en&tlng=en. doi:10.1590/S1516-31802012000100007.
- Shahlol AM, Abukhres OM, Taher IA. Prevalence and characterization of extended-spectrum β-lactamase-producing Enterobacteriaceae in Brack-Alshati, Fezzan, Libya. EC Microbiol. 2015;1(1):23-32. Available from: https://d1wqtxts1xzle7.cloudfront.net/91945792/ECMI-01-00004-libre.pdf?1664860941=.
- Ouchar Mahamat O, Kempf M, Lounnas M, Tidjani A, Hide M, Benavides JA, Carrière C, Bañuls AL, Jean-Pierre H, Ouedraogo AS, Dumont Y, Godreuil S. Epidemiology and prevalence of extended-spectrum β-lactamase- and carbapenemase-producing Enterobacteriaceae in humans, animals and the environment in West and Central Africa. Int J Antimicrob Agents. 2021;57(1):106203. Available from: https://linkinghub.elsevier.com/retrieve/pii/S092485792030409X. doi:10.1016/j.ijantimicag.2020.106203.
- Awulley Lelen Amina Emmanuela. Molecular epidemiology of extended-spectrum beta-lactamase (ESBL) producing Enterobacteriaceae from three tertiary hospitals in southern Ghana [master’s thesis]. Cape Coast: University of Cape Coast; 2018 Jul [cited 2025 Jul 21]. Available from: https://ir.ucc.edu.gh/xmlui/bitstream/handle/123456789/3841/EMMANUELA%2c%202018.pdf?sequence=1&isAllowed=y.
- Paterson DL, Bonomo RA. Extended-spectrum β-lactamases: a clinical update. Clin Microbiol Rev. 2005;18(4):657-86. Available from: https://journals.asm.org/doi/10.1128/CMR.18.4.657-686.2005. doi:10.1128/CMR.18.4.657-686.2005.
- Kaur S, Awari A. Burden of ESBL with antibiogram in clinical isolates of E. coli and Klebsiella species from tertiary care hospital in central India. Int J Med Res. 2018;5(4):497-503. Available from: https://ijmronline.org/article-details/8031. doi:10.18231/2394-5478.2018.0101.
- Sanou I, Kabore A, Tapsoba E, Bicaba I, Ba A, Zango B. Nosocomial urinary infections at the urology unit of the National University Hospital (Yalgado Ouedraogo), Ouagadougou: Feb.-Sept. 2012. Afr J Clin Exp Microbiol. 2015;16(1):1-6. Available from: https://www.ajol.info/index.php/ajcem/article/view/110715.
- Chaw PS, Schlinkmann KM, Raupach-Rosin H, Karch A, Pletz MW, Huebner J, Nyan O, Mikolajczyk R. Antibiotic use on paediatric inpatients in a teaching hospital in The Gambia, a retrospective study. Antimicrob Resist Infect Control. 2018;7:82. Available from: https://aricjournal.biomedcentral.com/articles/10.1186/s13756-018-0380-7. doi:10.1186/s13756-018-0380-7.
- Kebbeh A, Dsane-Aidoo P, Sanyang K, Darboe SM, Fofana N, Ameme D, Sanyang AM, Darboe KS, Darboe S, Sanneh B, Kenu E, Anto F. Antibiotics susceptibility patterns of uropathogenic bacteria: a cross-sectional analytic study at Kanifing General Hospital, The Gambia. BMC Infect Dis. 2023;23(1):723. Available from: https://bmcinfectdis.biomedcentral.com/articles/10.1186/s12879-023-08373-y. doi:10.1186/s12879-023-08373-y.
